Use a Laptop to Analyze 90% of Proteins, With a Single-GPU Inference Sequence Exceeding 10,000! Accelerate AlphaFold Inference by 5 Times and Reduce GPU Memory by 75%
Proteins are the basis of almost all functions of life. Evaluating the shape a protein folds — the “protein folding problem” — has been a notable challenge in biology for over 50 years. In response to this problem, AlphaFold introduced Transformer, a technology which achieves atomic-level accuracy in protein prediction, and is now widely used in biological research and drug development. Unfortunately, the actual deployment of the AlphaFold model brings many challenges:
- AlphaFold’s inference has a huge demand for GPU memory. Using the original AlphaFold to run inference of an amino acid sequence of 3,000 requires at least 60GB of GPU memory.
- The inference time for a long sequence takes several hours.
- Since MSA retrieval must be performed on several databases, preprocessing takes a lot of time.
Previously in March of this year, Colossal-AI’s open-source subsystem, FastFold, responded to these issues. Compared to AlphaFold, the speed on a single GPU was increased by 2.01 to 4.05 times. Now, a newer, and further improved version of FastFold is here!
The new version of FastFold reduces the 16GB memory required to infer protein structures of 1200 residues to just 5GB (single precision). This is done through fine-grain memory management optimization, which is much lower than the 8GB memory of the NVIDIA RTX3050. This means that users can use their own laptops to infer 90% of protein structures, significantly reducing deployment hardware costs! On a single NVIDIA A100 (80GB), FastFold increases the maximum amino acid sequence length for a single GPU to 10,000, covering 99.9999% of proteins. At the same time, FastFold is optimized by GPU Kernel using dynamic axis parallelism. The model inference speed is increased by 25% based on the optimization of the previous version, reaching an acceleration of 5 times. FastFold also uses full-process parallelization for pre-processing acceleration, significantly reducing the overall time for inference.
GitHub Repo:https://github.com/hpcaitech/ColossalAI#Biomedicine
Technical route
Fine-grained memory optimization
Although AlphaFold uses chunk technology, it is still very taxing on memory, a huge challenge for practical application. If we want to obtain a detailed structure for a 3000 amino acid protein, we need at least 60GB of memory. Only the most advanced professional graphics cards, (i.e. NVIDIA A100 80GB, worth about US$ 16, 000) can meet such memory requirements, discouraging many researchers and institutions.
FastFold reconstructs and optimizes AlphaFold’s chunk computing technology, making targeted adjustments to each module to optimize logic and efficiency. It also introduces local chunks of recalculation to reduce memory consumption of intermediate variables, expanding the chunk technology. With the help of chunk technology, the peak memory usage of the computing module is reduced by 40% during the model inference process.
For further improvement, we propose a memory sharing technology. By constructing a tensor variable structure during the inference process, each operation can modify the tensor in-place, avoiding copying memory during recursive function calls. Leveraging the memory sharing technology, memory consumption in the inference process can reduce the memory overhead by up to 50%.
With fine-grain memory management optimization, we are the first to perform inference of a 10,000 sequence protein, using half-precision bf16 on an A100, and to inference 7K amino acid sequences using single precision fp32.
In reality, 90% of the protein structure lengths are less than 1200 [1]. Through memory management technology, proteins with a sequence length of 1200 only need 5GB memory, which is much lower than the 8GB memory needed for NVIDIA RTX3050. This allows researchers to not need professional equipment. With the consumer-grade graphics card on your own laptop, you can get about 90% of the protein structure.

Chunk Strategy
GPU Kernel Optimization
FastFold uses computational optimization techniques such as operator fusion, and re-implements LayerNorm and Fused Softmax according to the performance characteristics of the AlphaFold model, which greatly improves the computational efficiency of AlphaFold on the GPU platform. The performance of the original AlphaFold is improved by 2 to 4 times, which greatly reduces the cost of deploying AlphaFold in actual scenarios. In the new version, we implemented a better operator based on Triton, and continued to achieve a speedup of about 25% on the basis of the original FastFold.
Dynamic axial parallelism
FastFold innovatively proposes dynamic axial parallelism technology based on the computational characteristics of AlphaFold. Unlike traditional tensor parallelism, dynamic axis-parallel selection divides data in the sequence direction of AlphaFold’s features and communicates using All_to_All. Dynamic axis parallelism has several advantages over tensor parallelism: 1) All computing modules in Evoformer are supported; 2) The amount of communication required is much smaller than that of tensor parallelism; 3) The memory consumption is lower than that of tensor parallelism; 4) DAP delivers further improvements on communication optimization, such as computing communication overlap. And we have added parallelization of ExtraMSA and TemplatePairStack in the new version. In actual deployment, dynamic axis parallelism can distribute computation to multiple GPUs, thereby greatly reducing the inference time of long-sequence models, and has a performance improvement of 9 to 11 times compared to the original AlphaFold (using 8 GPUs).
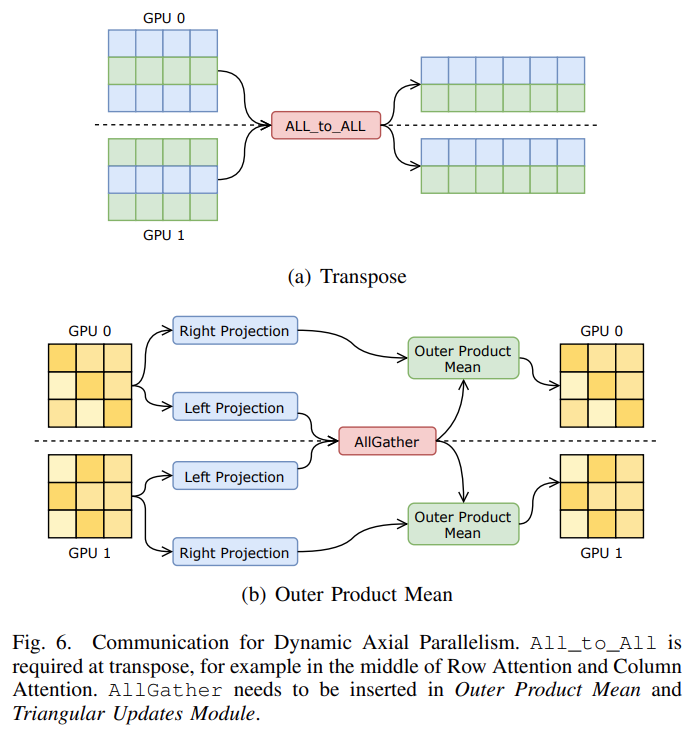
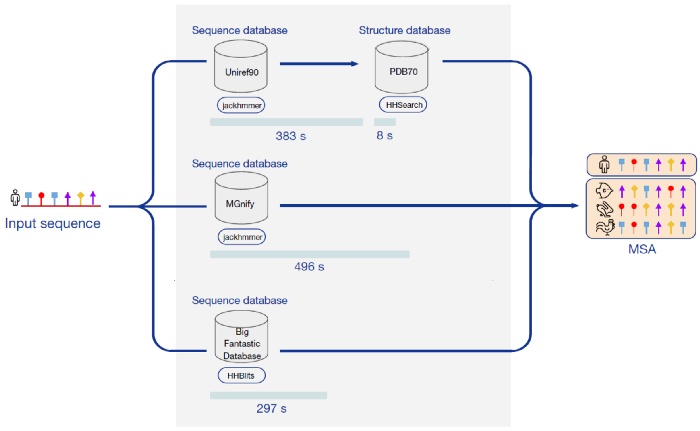
Parallel acceleration of the whole process
FastFold supports asynchronous distributed calls by using Ray as a distributed computing engine. Through Ray, we can find scheduling of various stages in the inference process of AlphaFold, and parallel the data preprocessing part. For protein structure of single amino acid sequences, we achieved about 3 times the preprocessing speedup. For protein structure composed of N amino acid sequences, we achieved about 3*N times the preprocessing speedup, which greatly reduced the deployment time and cost of AlphaFold.
Performance Testing
Memory test
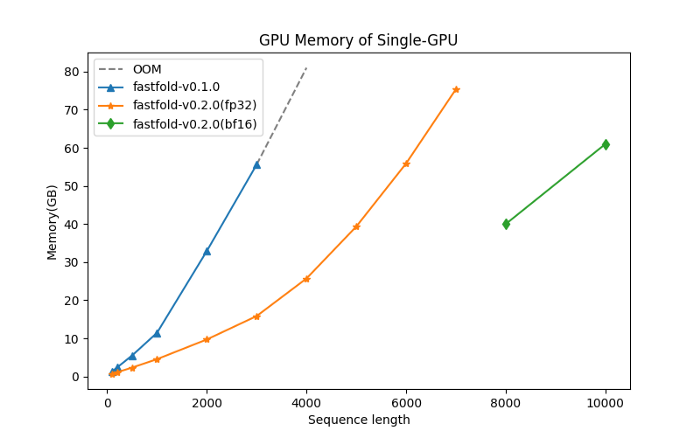
90% of the protein residues in nature are less than 1200. The new version of FastFold only needs 5GB of memory to inference the protein shape, and can be completed on a consumer-grade graphic card NVIDIA RTX3050.
The following figure shows the memory of FastFold when inferring sequences of different lengths in single precision (fp32). On a single NVIDIA A100 GPU (80GB), FastFold can efficiently inference up to 7k long amino acid sequences precisely. Using bf16, the maximum sequence length that can be found is increased to more than 10,000, a significant improvement.
Speed test
The following figure shows the time needed for FastFold to inference sequences of different lengths, as well as the effect of the number of GPUs on the inference speed with the sequence length as a constant at 2K. It can be seen that on the NVIDIA A100 GPU (80GB) hardware platform, the new version of FastFold single-card inference 2K sequence takes less than 10 minutes. If the hardware platform further expanded to 4*A100, dynamic axis parallelism would let FastFold complete inference in about 2.5 minutes, achieving an almost linear speedup.

The figure below shows the speed of the forward and backward propagation of FastFold’s single-layer Evoformer. Thanks to the further optimized GPU Kernel, the new version of FastFold’s single-layer Evoformer has an average speedup of about 25% compared to version 0.1. This means the new version of FastFold not only has excellent performance in inference, but also in training.

Conclusion
The proposed new version of FastFold aims at motivating the better application of protein structure prediction model training and inference in biomedicine and healthcare. By introducing fine-grain memory management optimization, the new version of FastFold is able to reduce the 16GB memory required to inference protein structures of 1200 residues to just 5GB (single precision). In addition, the new version of FastFold leverages the dynamic axial parallelism and full-process parallelization for pre-processing acceleration, making it possible to design and train larger models to get higher performance as well as significantly reducing the overall time for inference. According to our experiments, it is able to accelerate AlphaFold Inference by 5 times and reduce GPU memory by 75%. Further, an everyday laptop is enough to analyze 90% of the proteins, with singular-GPU inference sequence exceeding 10,000.
If you are interested, please check our project on GitHub: https://github.com/hpcaitech/ColossalAI#Biomedicine.
Reference:
[1] Tiessen, A., Pérez-Rodríguez, P. & Delaye-Arredondo, L.J. Mathematical modeling and comparison of protein size distribution in different plant, animal, fungal and microbial species reveals a negative correlation between protein size and protein number, thus providing insight into the evolution of proteomes. BMC Res Notes 5, 85 (2012). https://doi.org/10.1186/1756-0500-5-85
[2] Cheng, Shenggan, et al. “FastFold: Reducing AlphaFold Training Time from 11 Days to 67 Hours.” arXiv preprint arXiv:2203.00854 (2022).
Comments